Basic circadian methods
Behavpy_HMM is an extension of behavpy that revolves around the use of Hidden Markov Models (HMM) to segment and predict sleep and awake stages in Drosophila. A good introduction to HMMs is available here: https://web.stanford.edu/~jurafsky/slp3/A.pdf.
The provide a basic overview, HMMS are a generative probabilistic model, in which there is an assumption that a sequence of observable variables is generated by a sequence of internal hidden states. The transition probabilities between hidden states are assumed to follow the principles of a first order Markov chain.
Within this tutorial we will apply this framework to the analysis of sleep in Drosophila, using the observable variable movement to predict the hidden states: deep sleep, light sleep, quiet awake, active awake.
Hmmlearn
BehavpyHMM utilises the brilliant Python package Hmmlearn under the hood. Currently behavpy_HMM only uses the categorical version of the models available, as such it can only be trained and decode sequences that are categorical and in an integer form. If you wish to use their other models (multinomial, gaussian and guassian mixture models) or want to read more on their capabilities, head to their docs page for more information: https://hmmlearn.readthedocs.io/en/latest/index.html
Initialising behavpy_HMM
The HMM variant of behavpy is initialised in the same manner as the behavpy class. In fact, it is just a sub-classed version of behavpy so all the previous methods are still applicable, alongside the usual Pandas capabilities.
import ethoscopy as etho
# You can load you data using the previously mentioned functions
meta = etho.link_meta_index(meta_loc, remote, local)
data = etho.load_ethoscope(meta, min_time = 24, max_time = 48, reference_hour = 9.0)
# Remember you can save the data and metadata as pickle files, see the downloading section for a reminder about it
# to initialise a behavpy object, just call the class with the data and meta as arguments. Have check = True to ensure the ids match.
df = etho.behavpy_HMM(data, meta, check = True)Data curation
The data passed to the model needs to be good quality with long sequences containing minimal gaps and all a decent length in total.
Be careful when loading in your data. If you use the sleep_annotation() loading analysis function then it will automatically interpolate missing data as sleep giving you NaN (filler values) throughout your dataset. It's preferential to use the max_velocity_detector() function if you plan to use the HMM.
Use the .curate() and .bin_time() methods mentioned previously to curate the data. It is recommended to bin the data points to 60 second bins when analysing sleep and movement. But feel free to experiment with what works with your data.
import ethoscopy as etho
# load your data and metadata in and intialse your behavpy_HMM object
df = etho.behavpy_HMM(data, meta, check = True)
# it's often a good idea to filter out the start and beginning of experiments
df = df.t_filter(start_time = 24, end_time = 96)
# remove specimens with less than a days worth of data (if binned to 60 seconds)
df = df.curate(points = 1440)
# Any NaN values will throw an error when trying to train the HMM. It's good practice to check the data for any
# If below returns True you have NaN values in your data
print(np.any(np.isnan(df['YOUR COLUMN'].to_numpy())))
# If it is True then look at those values and replace or remove (the whole specimen) from the dataset
# Use below to view which rows have NaNs, see pandas documnetation for more information on how to replace NaN values
df[df['YOUR COLUMN'].isnull()]
# binning the variable can be done within the .hmm_train() method, see next!Training a HMM
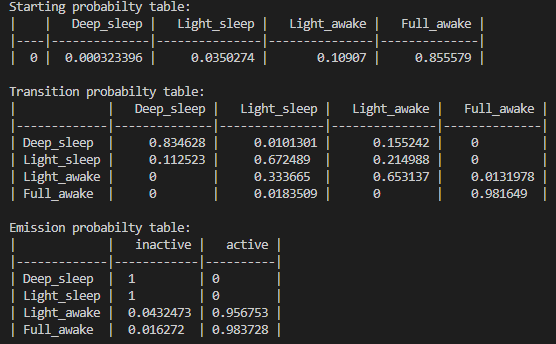
Behavpy_HMM provides a wrapper function for hmmlearn multinomial model. The method extends the hmmlearn model by automatically manipulating the data into a readable format for training. Additionally, the method provides the option to randomise the initialised transmission / observable parameters to train the model several times. Each iterative model is compared to the best previous performing model with the hopes of avoiding reach a local minima during hmmlearns own internal iterative process.As mentioned previously the HMM used here is categorical, therefore only select data that is categorically discrete. For example here we will be using activity, with not moving being one state and moving the other. These states as data must be in integer form. So this example, not moving will = 0 and moving will = 1. Follow this example if using more states, so if you have three states use the integers 0, 1, 2.In setting up the HMM you have to specify how many hidden states your system will have and the what states can transition into one another. See below for an example.
Setting up
# the setup for .hmm_train()
# name your hidden states and observables. This is just purely for visualising the output and setting the number of hidden states!
hidden_states = ['Deep sleep', 'Light sleep', 'Light awake', 'Active awake']
observable_variables = ['inactive', 'active']
# if you believe your hidden states can only transition into specific states or there's a flow you can setup the model to train with this in mind. Have the transitions you want to happen set as 'rand' and those that can't as 0
t_prob = np.array([['rand', 'rand', 'rand', 0.00],
['rand', 'rand', 'rand', 0.00],
[0.00, 'rand', 'rand', 'rand'],
[0.00, 'rand', 'rand', 'rand']])
# Make sure your emission matrix aligns with your categories
em_prob = np.array([[1, 0],
[1,0],
['rand', 'rand'],
['rand', 'rand']])
# the shape of each array should be len(hidden_states) X len(names). E.g. 4x4 and 4x2 respectivelyYou can leave trans_probs and em_prob as None to have all transitions randomised before each iteration
Training
# add the above variables to the hmm_train method
# iterations is the number of loops with a new randomised parameters
# hmm_interactions is the number of loops within hmmlearn before it stops. This is superceeded by tol, with is the difference in the logliklihood score per interation within hmmlearn
# save the best performing model as a .pkl file
df.hmm_train(
states = hidden_states,
observables = observable_variables,
var_column = 'moving',
trans_probs = t_prob,
emiss_probs = em_prob,
start_probs = None,
iterations = 10,
hmm_iterations = 100,
tol = 2,
t_column = 't',
bin_time = 60,
file_name = 'experiment_hmm.pkl', # replace with your own file name
verbose = False)
# If verbose is true the logliklihood per hmm iteraition is printed to screenDiving deeper! To find the best model the dataset will be split into a test (10%) and train (90%) datasets. Each successive model will be scored against the best on the test dataset. The best scoring model will be the final save to your file name.